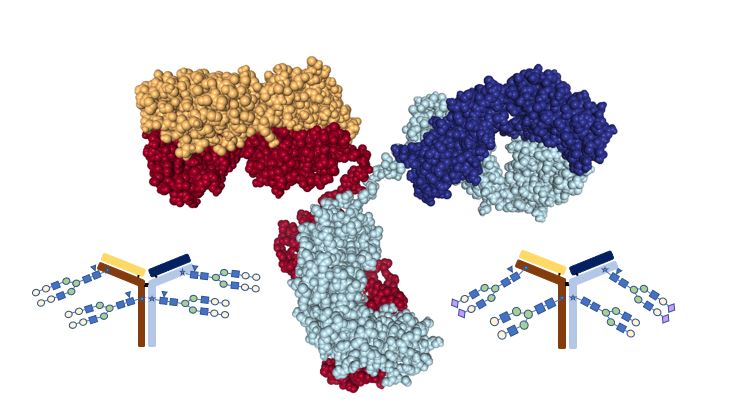
Antibodies are proteins. In the body, they are made by B cells of the immune system. To produce antibodies as therapeutics, they are synthesized by cells in culture. The structure of an antibody is defined by the amino acid sequence of the protein (Figure 1). The incredible diversity of antibodies, which is essential to our ability to fight pathogens, comes from having many genes encoding the peptides that come together to make a functional antibody. Additional variation comes from rearrangement of the genes in what are called hypervariable regions. Antibodies are not only composed of amino acids though. The cells that produce them also add sugars to them to form complex long chains. These sugar modifications are called glycosylation and produce yet another layer of functional diversity in antibodies, even antibodies with the same amino acid sequence.
Because cells make antibodies, even those that will be used as medicine, the composition of the final product is difficult to control. Even using molecular biology and genetic engineering techniques to ensure that the antibody always has the same amino acid sequence does not ensure that every antibody in the preparation will be identical or that every batch of cultured cells will make a consistent amount of stable, properly folded antibody protein. Add to that the impossibility of controlling the sugar modifications and the final product is always going to be a mixture of glycosylated antibodies with the same amino acid sequence even when produced by the same supplier using the same conditions.
The labs of Lai-Xi Wang from the University of Maryland, College Park and Jeffrey Ravetch from The Rockefeller University have developed a technique for controlling the sugar chains that are attached to antibody-based drugs. They applied this new process to an antibody-based drug used to treat cancers that depend on the growth factor receptor called EGFR. The drug is cetuximab (brand name Erbitux), which is approved for use in treating patients with metastatic head and neck cancers and metastatic colorectal cancers. This is an important advance for developing improved versions of cetuximab and the development and improvement of other antibody-based drugs.
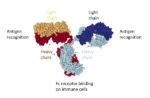
Antibodies have two main parts (Figure 2): The Fab part of the antibody, which binds to the antigen, in this case a growth factor receptor, and the Fc part of the antibody, which binds to immune cells. As the cells make the antibody, complex chains of sugar molecules are added to the different parts of the antibody through a process called glycosylation. Glycosylation patterns can be highly variable and can affect how an antibody functions. Some glycosylation patterns reduce the ability of an antibody to stimulate a cytotoxic response, which is necessary for an anti-tumor antibody to be effective. Other patterns, including a common one on cetuximab, can trigger severe or fatal allergic reactions in some patients. Ideally, drug companies would like to be able to control exactly which sugar chains are present on the Fab part and which are present on the Fc part. This has not been possible until now.
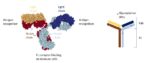
Using a process called chemoenzymatic modification, the researchers took commercially available preparations of cetuximab, which had been made by live cells in culture and had diverse glycosylation patterns, and incubated the cetuximab with bacterial enzymes that removed all but the first sugar molecules (core glycosylation) attached to the Fab and Fc parts of the antibodies. These core-glycosylated antibodies were the exposed in a specific order with other bacterial enzymes that added complex sugars to or removed complex sugars from either the sugar chains attached to the Fab or the Fc parts of the antibodies. In this way, the researchers remodeled the glycosylation pattern of cetuximab.
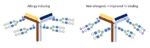
One way that cetuximab treats cancer is by helping immune cells find and kill the cancer cells. The remodeling of the Fc parts of cetuximab antibodies made the antibodies bind more strongly to the immune cell receptor. The remodeled cetuximab were better at promoting immune cells to kill cancer cells in culture. Another advantage of the glycan-remodeled cetuximab was the elimination of the allergic sugars in the commercial preparation. The modified cetuximab would not trigger the most commonly occurring type allergic reaction that many patients have with this antibody-based treatment. Thus, by remodeling the sugar chains, the researchers made a safer and more effective version of cetuximab (Figure 3).
The key to the approach is the bacterial enzymes that recognize and specifically modify the sugar chains. The researchers used a combination of naturally occurring enzymes from bacteria and enzymes that had been engineered by introducing point mutations to alter their specificity. As Wang states, “Enzyme substrate specificity is the name of the game.” This study provides a method for improving antibody-based drugs, which are widely used for treating cancer and autoimmune diseases.
In related commentary, I describe research into optimizing therapeutic antibodies by modifying the amino acid sequence of the heavy chains to alter the immune cells that the Fc portion binds and to improve tumor-killing efficacy.
Related Resources
J. P. Giddens, J. V. Lomino, D. J. DiLillo, J. V. Ravetch, L.-X. Wang, Site-selective chemoenzymatic glycoengineering of Fab and Fc glycans of a therapeutic antibody. Proc. Natl. Acad. Sci. U. S. A. (2018). DOI: 10.1073/pnas.1812833115 Online Text
N. R. Gough, Optimizing anti-tumor therapies with engineered antibodies. BioSerendipity (18 September 2018) https://www.bioserendipity.com/optimizing-anti-tumor-therapies-with-engineered-antibodies/
Ravetch Laboratory, The Rockefeller University: https://www.rockefeller.edu/our-scientists/heads-of-laboratories/889-jeffrey-v-ravetch/(accessed 5 September 2018)
Wang Group, University of Maryland, College Park: http://glycobio.wixsite.com/wangglycan (accessed 5 November 2018)
Cite this article:
N. R. Gough, Building Better Antibodies. BioSerendipity (6 November 2018) https://www.bioserendipity.com/building-better-antibodies/.
Also of Interest in BioSerendipity