Researchers commonly refer to the molecular and biochemical events that occur in response to extracellular signals as pathways. There is a tendency to lump a related set of pathways into a general overarching pathway. For example, researchers may refer to the TGF-β pathway or the Wnt pathway. These are no more individual pathways than the fish in the sea are all the same kind. Yes, they have elements in common, just as fish have mouths, fins, and scales. Yet, these pathways represent a collection of functionally related events or molecular processes mediated by structurally related proteins. This reliance on shorthand makes it easier to convey general concepts but glosses over the details in a way that can be either misinterpreted or unintentionally mislead the reader. When a researcher uses the published literature to guide future experiments or seek funding, lack of precision in understanding exactly what the author meant by “pathway” can lead to wasted resources, an apparent inability to reproduce the published work, and frustration. When a reviewer encounters this imprecision, it can lead to rejection of the submitted manuscript or a poor score for a research grant, even when the data or proposed studies are important and advance the field.
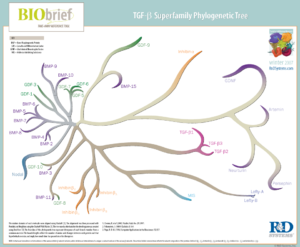
The TGF-β and Wnt pathways illustrate this semantic problem. In humans, the ligands of the TGF-β family are so numerous that they are often grouped on the basis of sequence similarity (Figure 1). For example, there is a group with the name TGF-β, a group with the name BMP, and a group named GDF. The TGF-β superfamily signals through a family of receptors, which are organized into two groups, the type I and the type II receptors (Figure 2).
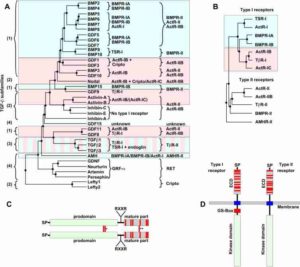
Each functional receptor consists of one protein from each group and these dimeric receptor complexes can interact with a coreceptor protein. Frequently, researchers will refer to the transcriptional regulatory events mediated by the Smad proteins (SMAD1, 2, 3, 4, 5, or 8), which are regulated by the activity of the receptors, as the “canonical TGF-β pathway” without specifying exactly which ligand, receptor, and Smad complex perform the signaling. Furthermore, changes in gene expression mediated by receptor-activated Smads are only one of the many signaling events that can be triggered by signaling through this family of ligands and receptors. Consequently, referring to the “TGF-β Pathway” is not precise and typically encompasses only the Smad-mediated transcriptional regulatory output of this diverse set of ligands and receptors. Having to write the “TGFB1-TGFBRI/TGFBRII-SMAD2/SMAD4” pathway every time takes up a lot of space and precious words (especially when word count or pages are limited) and makes the text hard to follow, especially if this long phrase is frequently repeated. It is easy to see how the author may revert to the simple shorthand TGF-β pathway or BMP pathway. However, it can become very confusing when an author refers to the entire superfamily of ligands and receptors and the downstream signaling events as the TGF-β pathway. In this case, the reader is left wondering if the author means the TGF-β superfamily of pathways, a subset of pathways mediated by the TGF-β subfamily of ligands, or a specific pathway mediated by a specific individual member of the TGF-β subfamily of ligands, a specific receptor complex, and specific downstream mediators.
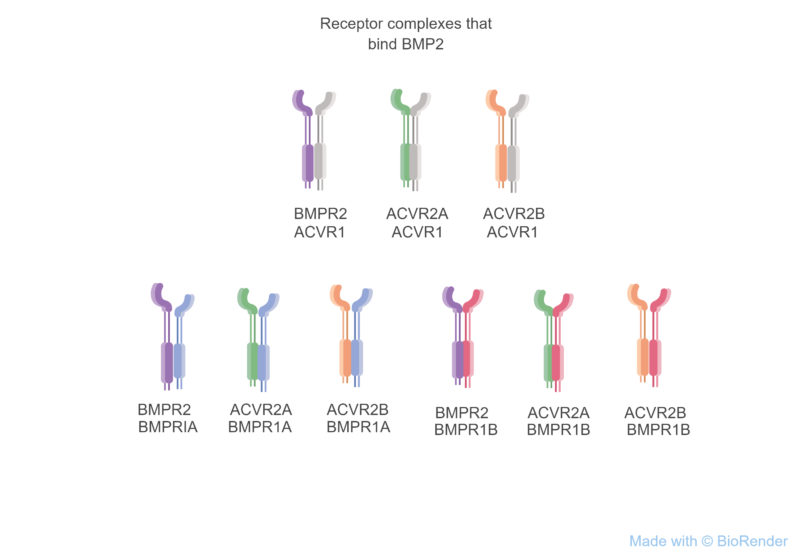
Although the mapping of ligand subfamilies to subsets of receptor complexes and Smad mediators for the TGF-β pathway is fairly well established, as illustrated by the 9 receptor complexes that bind BMP2 (Figure 3), this is not the case for the Wnt pathway. In humans, the Wnt pathway has 19 genes encoding ligands and 15 receptor- or coreceptor-encoding genes. Exactly which ligands activate each receptor or receptor complex in each cell is still an area of active investigation, although advances in gene knockout using CRISPR technology may provide rapid progress. Current data indicate that many of the ligands are promiscuous and can activate the Wnt-dependent transcriptional pathway through one of several of the receptors, and a few ligands are highly selective in the receptors that they bind and activate. Most of the studies have focused on the transcriptional output of Wnt signaling, which is mediated through stabilization of the protein β-catenin. Research into the β-catenin-independent transcriptional and nontranscriptional outputs of Wnt signaling lags behind, leaving an opportunity for this gap in knowledge of how specific Wnt ligands interact with specific receptor complexes mediate these other cellular responses.
Even relatively simple signaling pathways with small numbers of ligands and receptors, such as the EGF pathway or adrenergic signaling, can be complex. In humans, the EGF family of ligands has 10 members, but there are only 4 receptors. Compared to the Wnt or TGF-β pathways, this seems relatively simple. However, the ligands form hetero and homodimers, the receptors dimerize, and the downstream mediators are diverse. Even adrenergic signaling, which only involves two natural ligands, adrenaline (also known as epinephrine) and noradrenaline (also known as norepinephrine), involves 9 different receptors of the G protein-coupled receptor (GPCR) family.
Where does that leave the author and reader? My recommendation, which is based on 17 years of experience as an editor for a signaling journal and signaling database, is to use the shorthand when introducing the study or research (the 10,000 foot view). When discussing the data, be as precise as possible. If a shorthand terminology is used to refer to a particular ligand-receptor-mediator pathway, be sure to explicitly and precisely define the shorthand early in the article or grant. When the exact molecules participating in the signaling event are unknown, be sure it is clearly stated what details are unknown. Avoid vague, ambiguous language. Let the reader know what the limits of the study are so that future work can fill in the gaps. Precision and accuracy, along with clearly stating the limits, improves reproducibility in science and prevents readers from being unintentionally mislead and can increase the chance of passing in-depth review.
Related Reading
O. Voloshanenko, P. Gmach, J. Winter, D. Kranz, M. Boutros, Mapping of Wnt-Frizzled interactions by multiple CRISPR targeting of receptor gene families. FASEB J. 31, 4832-4844 (2017). PubMed
J. P. Dijksterhuis, B. Baljinnyam, K. Stanger, H. O. Sercan, Y. Ji, O. Andres, J. S. Rubin, R. N. Hannoush, G. Schulte, Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290, 6789-6798 (2015). PubMed
T. D. Meuller, J. Nickel, Promiscuity and specificity in BMP receptor activation. FEBS Lett. 586, 1846-1859 (2012). PubMed
Genes Encoding Ligands of the TGF-β Superfamily in Humans
TGFB1, TGFB2, TGFB3, BMP2, BMP3, BMP4, BMP5, BMP6, BMP7, BMP8A, BMP8B, BMP10, BMP15, GDF1, GDF2, GDF3, GDF5, GDF6, GDF7, GDF9, GDF10, GDF15, INHBA, INHBB, INHBC, INHA, INHBE, NODAL, LEFTY2, MSTN, AMH
Genes Encoding TGF-β Type I and Type II Receptors and Coreceptors
TGFBR1, ACVRL1, ACVR1, ACVR1B, ACVR1C, BMPR1A, BMPR1B, TGFBR2, ACVR2A, ACVR2B, AMHR2, BMPR2, TGFBR3, ENG
Genes Encoding Wnt Ligands in Humans
WNT1, WNT2, WNT2B, WNT3, WNT3A, WNT4, WNT5A, WNT5B, WNT7A, WNT7B, WNT8A, WNT8B, WNT9A, WNT9B, WNT10A, WNT10B, WNT11, WNT16
Genes Encoding Wnt Receptors and Coreceptors in Humans
FZD1, FZD2, FZD3, FZD4, FZD5, FZD6, FZD7, FZD8, FZD9, FZD10, ROR1, ROR2, RYK, LRP5, LRP6
Cite as: N. R. Gough, TGF-β and Wnt Signaling Challenge the Concept of ‘a’ Pathway. BioSerendipity (12 March 2018) https://www.bioserendipity.com/tgf-%CE%B2-and-wnt-signaling-challenge-the-concept-of-a-pathway/