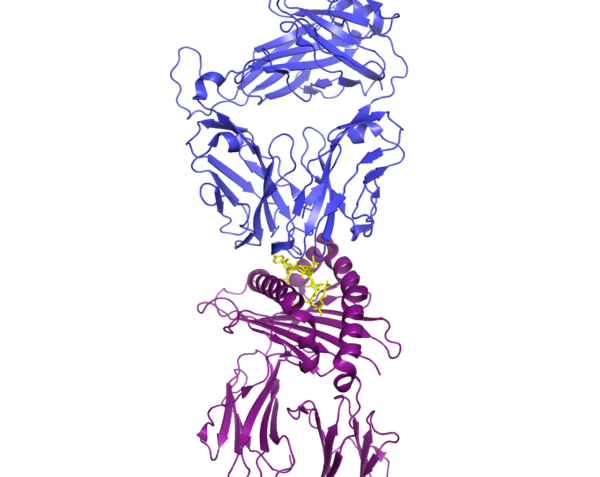
T cells are a major component of the adaptive immune system. The video shows the intracellular events that let T cells learn about and recognize virus-infected cells.
Infected cells can use the major histocompatibility complex (MHC) I or MHC II pathway to signal to T cells that they are infected. Both pathways rely on the degradation of viral proteins into peptides that can be displayed on the cell surface to enable viral recognition. Lysosomes or intracellular compartments that are similar to lysosomes and contain protein-degrading enzymes play a critical role in the MHC II pathway. A cellular machine called the proteasome that breaks down proteins in the cytosol plays a critical role in the MHC I pathway. Through these two pathways, T cells can recognize infected cells through receptor complexes that specifically bind to individual fragments of the viral proteins. Thus, most viruses cannot mutate so much that all of the fragments that could be recognized by T cells will be eliminated. Thus, a little immune training goes a long way. The video shows how these two cellular process work to enable two types of T cells to recognize infected cells using the virus that causes COVID-19 as an example.
In humans the MHC proteins are referred to as human leukocyte antigens (HLAs). The genes encoding these proteins are highly polymorphic, meaning that they are variable across the population. Combined with the highly variable T cell receptors, the two MHC pathways let an individual’s immune system tell the difference between tell their own cells and someone else’s and between their own healthy cells and those that have been infected. Differences in HLA-encoding genes are associated with whether an individual is likely to develop symptomatic or asymptomatic COVID-19.
Similar to recognition of the virus upon cellular infection, vaccines leverage the same pathways to display the antigens contained in or encoded by the vaccine. For vaccines, the display of the antigens occurs primarily through antigen-presenting cells of the immune system rather than virally infected cells. Vaccines that will have the broadest efficacy across populations will include epitopes that are recognized by most individuals regardless of the variation in their HLA-encoding genes.
References
Augusto DG, Yusufali T, Peyser ND, Butcher X, Marcus GM, Olgin JE, Pletcher MJ, Maiers M, Hollenbach JA. HLA-B*15:01 is associated with asymptomatic SARS-CoV-2 infection. medRxiv [Preprint]. (2021 Sep 10) 2021.05.13.21257065. DOI: 10.1101/2021.05.13.21257065.
Cun Y, Li C, Shi L, Sun M, Dai S, Sun L, Shi L, Yao Y. COVID-19 coronavirus vaccine T cell epitope prediction analysis based on distributions of HLA class I loci (HLA-A, -B, -C) across global populations. Hum. Vaccin. Immunother. 17, 1097-1108 (2021). DOI: 10.1080/21645515.2020.1823777.
Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J. Biol. Chem. 295, 12910-12934 (2020). DOI: 10.1074/jbc.REV120.013930.
He J, Huang F, Zhang J, Chen Q, Zheng Z, Zhou Q, Chen D, Li J, Chen J. Vaccine design based on 16 epitopes of SARS-CoV-2 spike protein. J. Med. Virol. 93, 2115-2131 (2021). DOI: 10.1002/jmv.26596.
Lubin JH, Zardecki C, Dolan EM, Lu C, Shen Z, Dutta S, Westbrook JD, Hudson BP, Goodsell DS, Williams JK, Voigt M, Sarma V, Xie L, Venkatachalam T, Arnold S, Alfaro Alvarado LH, Catalfano K, Khan A, McCarthy E, Staggers S, Tinsley B, Trudeau A, Singh J, Whitmore L, Zheng H, Benedek M, Currier J, Dresel M, Duvvuru A, Dyszel B, Fingar E, Hennen EM, Kirsch M, Khan AA, Labrie-Cleary C, Laporte S, Lenkeit E, Martin K, Orellana M, Ortiz-Alvarez de la Campa M, Paredes I, Wheeler B, Rupert A, Sam A, See K, Soto Zapata S, Craig PA, Hall BL, Jiang J, Koeppe JR, Mills SA, Pikaart MJ, Roberts R, Bromberg Y, Hoyer JS, Duffy S, Tischfield J, Ruiz FX, Arnold E, Baum J, Sandberg J, Brannigan G, Khare SD, Burley SK. Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first 6 months of the COVID-19 pandemic. Proteins 90, 1054-1080 (2022). DOI: 10.1002/prot.26250.
Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell. Biol. 23, 21-39 (2022). DOI: 10.1038/s41580-021-00432-z.
Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203-216 (2015). DOI: 10.1038/nri3818.
van de Leemput J, Han Z. Understanding individual SARS-CoV-2 Proteins for Targeted Drug Development against COVID-19. Mol. Cell. Biol. 41, e0018521 (2021). https://doi.org/10.1128/MCB.00185-21
Also of interest
How Killer and Helper T Cells Recognize Viruses
More about COVID-19 by Nancy R. Gough
Cite as: N. R. Gough, Two pathways to viral recognition. BioSerendipity (29 December 2022) https://www.bioserendipity.com/two-pathways-to-viral-recognition/