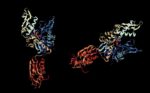
In a study of infection by bacteria of the mycoplasma family, researchers at the University of Maryland School of Medicine have discovered that the bacterial protein DnaK triggers a cascade of events that can cause infected cells and nearby uninfected cells to become cancerous. DnaK is a prokaryotic stress-induced protein (Figure 1) that is a member of the large family of such proteins referred to as heat shock proteins or chaperones. In bacteria, DnaK interacts with many proteins to help them properly fold or function or to protect them from damage during periods of environmental stress, such as exposure to higher than optimal temperatures.
There are many strains of mycoplasma. Some strains have been associated with cancers that occur in people infected with HIV, the virus that causes AIDS. Like many cancers, these mycoplasma-associated cancers have impaired function of a key cellular protein called p53 that helps cells cope with DNA damage. When p53 is not functioning properly, DNA damage tends to accumulate and cells with damage DNA proceed to divide, even though they should not. Two of the body’s main protective mechanisms against cancer are p53-mediated cell cycle arrest to enable time for DNA repair and a form of cell death called apoptosis. Apoptosis is triggered by p53 in cells with damaged DNA that cannot be or has not been repaired. Thus, not only do cells with impaired p53 not respond properly to DNA damage, instead of dying they keep dividing. This is a recipe for cancer. Indeed, more than half of human cancers have mutations or dysfunctional p53.
By searching for mycoplasma proteins that interacted directly or indirectly with p53, Zella and colleagues identified DnaK as one of the bacterial proteins that could be isolated with p53 from infected cells. Furthermore, the infected cells had impaired p53 function and were resistant to two drugs used as anticancer treatments, nutlin and 5-fluorouracil (5-FU). Both of these drugs kill cancer cells, but they require p53 for effectiveness.
DnaK did not appear to directly bind p53, so the researchers performed an experiment to identify proteins that were isolated with DnaK, instead of p53, from infected cells. Three cellular proteins were identified that directly interacted with DnaK: USP10, PARP1, and DNAJA1.
The interaction between DnaK and USP10 revealed how DnaK reduced p53 function. USP10 modifies p53 so that it does not get degraded. DnaK blocked this activity of USP10, leading to degradation of p53. This explained why infected cells were resistant to the death-inducing effects of nutlin and 5-FU.
The interaction between DnaK and PARP1 impaired PARP1 activity. PARP1 is a critical protein needed to repair DNA damage. Thus, by inhibiting PARP1, DnaK would trigger genome instability.
The interaction with DNAJ1A1 could provide a mechanism for DnaK to interfere with the folding of many cellular proteins. In the bacteria, DnaK needs two other proteins to perform its chaperone function. DNAJA1 is the human protein that is similar to one of these bacterial DnaK helper proteins and could substitute for the bacterial versions in promoting the enzyme activity of DnaK that is part of its chaperone function. DNAJA1 interacts with proteins at mitochondria. Consequently, by interacting with DNAJA1, DnaK may affect mitochondrial function or the involvement of mitochondria in apoptosis.
The researchers did not detect much bacterial DNA in the cancers, suggesting that the infection did not have to persist for cancer to develop. Indeed, cells did not even have to be infected with the bacteria for them to acquire DnaK. Cells exposed to DnaK in the medium took up this protein. Thus, cells near the infected cells could become cancer by taking up DnaK released their infected neighbors. The scientists called this a “hit-and-run” mechanism of oncogenesis.
Other bacteria might contribute to cancer through a similar mechanism involving DnaK. Indeed, comparing the amino acid sequence of these proteins from different species of bacteria revealed that the bacteria associated with cancers had DnaK proteins that were similar to each other. The DnaK proteins from bacteria that are not associated with cancers are different. This study suggests that infections with bacteria with this cancer-associated form of DnaK may contribute to many more cancers than previously thought.
Highlighted Article
D. Zella, S. Curreli, F. Benedetti, S. Krishnan, F. Cocchi, O. S. Latinovic, F. Denaro, F. Romerio, M. Djavani, M. E. Charurat, J. L. Bryant, H. Tettelin, R. C. Gallo, Mycoplasma promotes malignant transformation in vivo, and its DnaK, a bacterial chaperon protein, has broad oncogenic properties. Proc. Natl. Acad. Sci. U. S. A. (3 December 2018) DOI: 10.1073/pnas.1815660115 Full Text
Related Resources
Chaperone DnaK (IPR012725), InterPro. EMBL-EBI http://www.ebi.ac.uk/interpro/entry/IPR012725 (accessed 27 November 2018).
Structure of DnaK: Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with ADP and Pi. DOI: 10.2210/pdb5OBV/pdb
TUMOR PROTEIN p53, OMIM. https://www.omim.org/entry/191170 (accessed 27 November 2018).
TP53, UniProt. https://www.uniprot.org/uniprot/P04637 (accessed 27 November 2018).
DNAJA1, Uniprot. https://www.uniprot.org/uniprot/P31689 (accessed 27 November 2018)
About Dr. Robert C. Gallo, Institute of Human. Virology http://www.ihv.org/about/About-Dr-Robert-C-Gallo/ (accessed 4 December 2018)
Cite as: N. R. Gough, Bacterial Chaperone Protein Causes Cancer. BioSerendipity (7 December 2018) https://www.bioserendipity.com/bacterial-chaperone-protein-causes-cancer.